36 混合效应模型
\[ \def\bm#1{{\boldsymbol #1}} \]
I think that the formula language does allow expressions with ‘/’ to represent nested factors but I can’t check right now as there is a fire in the building where my office is located. I prefer to simply write nested factors as
factor1 + factor1:factor2.— Douglas Bates 1
混合效应模型在心理学、生态学、计量经济学和空间统计学等领域应用十分广泛。线性混合效应模型有多个化身,比如生态学里的分层线性模型(Hierarchical linear Model,简称 HLM),心理学的多水平线性模型(Multilevel Linear Model)。模型名称的多样性正说明它应用的广泛性! 混合效应模型内容非常多,非常复杂,因此,本章仅对常见的四种类型提供入门级的实战内容。从频率派和贝叶斯派两个角度介绍模型结构及说明、R 代码或 Stan 代码实现及输出结果解释。
除了 R 语言社区提供的开源扩展包,商业的软件有 Mplus 、ASReml 和 SAS 等,而开源的软件 OpenBUGS 、 JAGS 和 Stan 等。混合效应模型的种类非常多,一部分可以在一些流行的 R 包能力范围内解决,其余可以放在更加灵活、扩展性强的框架 Stan 内解决。因此,本章同时介绍 Stan 框架和一些 R 包。
本章用到 4 个数据集,其中 sleepstudy 和 cbpp 均来自 lme4 包 (Bates 等 2015),分别用于介绍线性混合效应模型和广义线性混合效应模型,Loblolly 来自 datasets 包,用于介绍非线性混合效应模型,最后,朗格拉普岛核辐射数据数据集用于介绍广义可加混合效应模型。
在介绍理论的同时给出 R 语言或 S 语言实现的几本参考书籍。
- 《Mixed-Effects Models in S and S-PLUS》(Pinheiro 和 Bates 2000)
- 《Mixed Models: Theory and Applications with R》(Demidenko 2013)
- 《Linear Mixed-Effects Models Using R: A Step-by-Step Approach》(Gałecki 和 Burzykowski 2013)
- 《Linear and Generalized Linear Mixed Models and Their Applications》(Jiang 和 Nguyen 2021)
36.1 线性混合效应模型
I think what we are seeking is the marginal variance-covariance matrix of the parameter estimators (marginal with respect to the random effects random variable, B), which would have the form of the inverse of the crossproduct of a \((q+p)\) by \(p\) matrix composed of the vertical concatenation of \(-L^{-1}RZXRX^{-1}\) and \(RX^{-1}\). (Note: You do not want to calculate the first term by inverting \(L\), use
solve(L, RZX, system = "L")
[…] don’t even think about using
solve(L)don’t!, don’t!, don’t!
have I made myself clear?
don’t do that (and we all know that someone will do exactly that for a very large \(L\) and then send out messages about “R is SOOOOO SLOOOOW!!!!” :-) )
— Douglas Bates 2
- 一般的模型结构和假设
- 一般的模型表达公式
-
nlme 包的函数
lme() - 公式语法和示例模型表示
线性混合效应模型(Linear Mixed Models or Linear Mixed-Effects Models,简称 LME 或 LMM),介绍模型的基础理论,包括一般形式,矩阵表示,参数估计,假设检验,模型诊断,模型评估。参数方法主要是极大似然估计和限制极大似然估计。一般形式如下:
\[ \bm{y} = X\bm{\beta} + Z\bm{u} + \bm{\epsilon} \]
其中,\(\bm{y}\) 是一个向量,代表响应变量,\(X\) 代表固定效应对应的设计矩阵,\(\bm{\beta}\) 是一个参数向量,代表固定效应对应的回归系数,\(Z\) 代表随机效应对应的设计矩阵,\(\bm{u}\) 是一个参数向量,代表随机效应对应的回归系数,\(\bm{\epsilon}\) 表示残差向量。
一般假定随机向量 \(\bm{u}\) 服从多元正态分布,这是无条件分布,随机向量 \(\bm{y}|\bm{u}\) 服从多元正态分布,这是条件分布。
\[ \begin{aligned} \bm{u} &\sim \mathcal{N}(0,\Sigma) \\ \bm{y}|\bm{u} &\sim \mathcal{N}(X\bm{\beta} + Z\bm{u},\sigma^2W) \end{aligned} \]
其中,方差协方差矩阵 \(\Sigma\) 必须是半正定的,\(W\) 是一个对角矩阵。nlme 和 lme4 等 R 包共用一套表示随机效应的公式语法。
sleepstudy 数据集来自 lme4 包,是一个睡眠研究项目的实验数据。实验对象都是有失眠情况的人,有的人有严重的失眠问题(一天只有 3 个小时的睡眠时间)。进入实验后的前10 天的情况,记录平均反应时间、睡眠不足的天数。
#> 'data.frame': 180 obs. of 3 variables:
#> $ Reaction: num 250 259 251 321 357 ...
#> $ Days : num 0 1 2 3 4 5 6 7 8 9 ...
#> $ Subject : Factor w/ 18 levels "308","309","310",..: 1 1 1 1 1 1 1 1 1 1 ...Reaction 表示平均反应时间(毫秒),数值型,Days 表示进入实验后的第几天,数值型,Subject 表示参与实验的个体编号,因子型。
#> Subject
#> Days 308 309 310 330 331 332 333 334 335 337 349 350 351 352 369 370 371 372
#> 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> 3 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> 4 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> 5 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> 6 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> 7 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> 8 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> 9 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1每个个体每天产生一条数据,下 图 36.1 中每条折线代表一个个体。
library(ggplot2)
ggplot(data = sleepstudy, aes(x = Days, y = Reaction, group = Subject)) +
geom_line() +
scale_x_continuous(n.breaks = 6) +
theme_bw() +
labs(x = "睡眠不足的天数", y = "平均反应时间")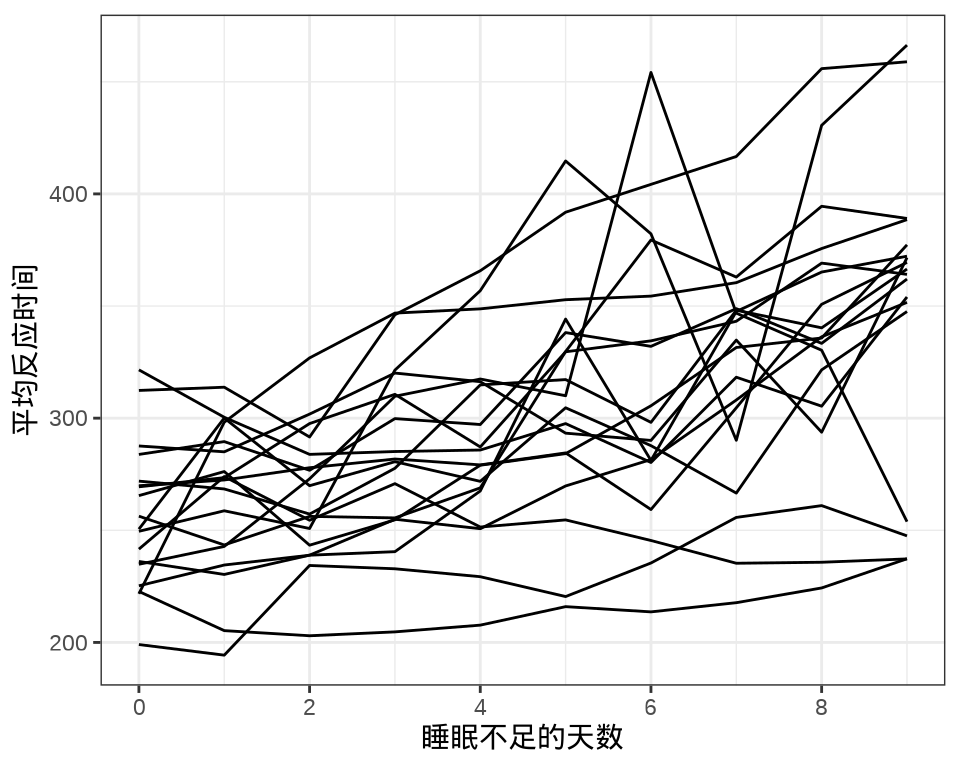
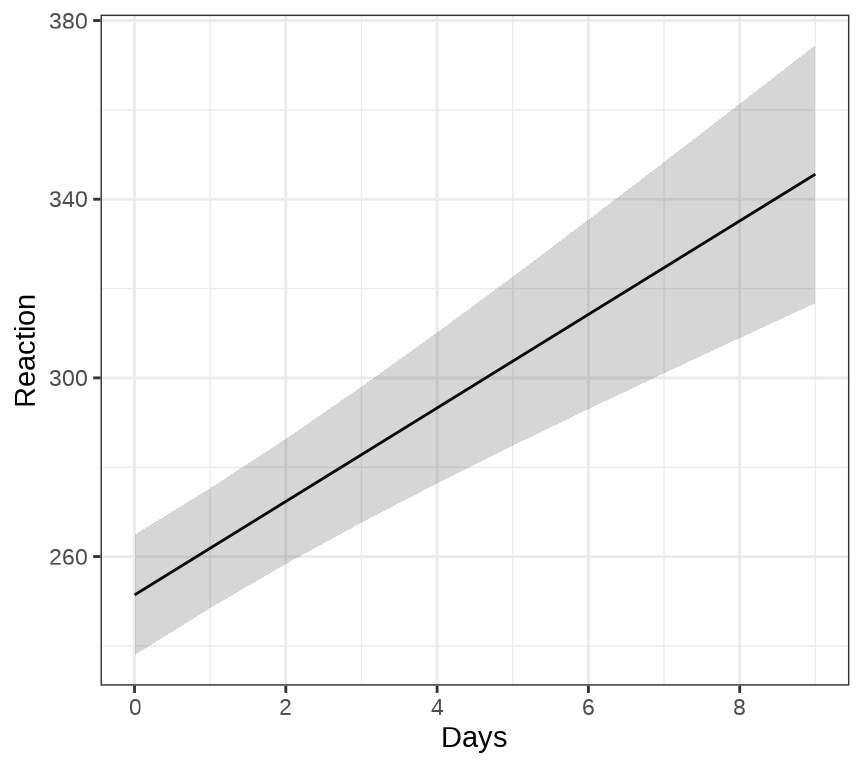
对于连续重复测量的数据(continuous repeated measurement outcomes),也叫纵向数据(longitudinal data),针对不同个体 Subject,相比于上图,下面绘制反应时间 Reaction 随睡眠时间 Days 的变化趋势更合适。图中趋势线是简单线性回归的结果,分面展示不同个体Subject 之间对比。
ggplot(data = sleepstudy, aes(x = Days, y = Reaction)) +
geom_point() +
geom_smooth(formula = "y ~ x", method = "lm", se = FALSE) +
scale_x_continuous(n.breaks = 6) +
theme_bw() +
facet_wrap(facets = ~Subject, labeller = "label_both", ncol = 6) +
labs(x = "睡眠不足的天数", y = "平均反应时间")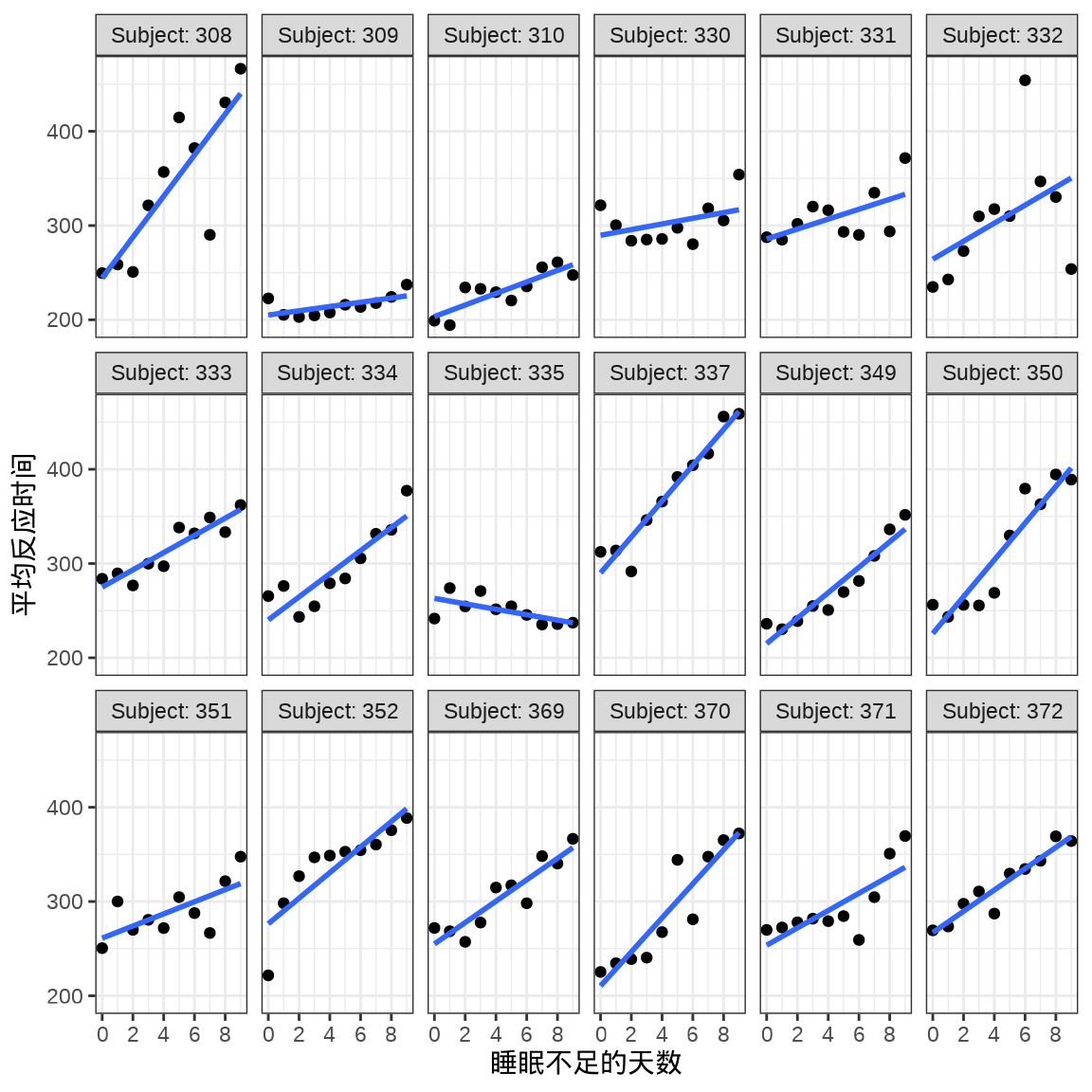
36.1.1 nlme
考虑两水平的混合效应模型,其中随机截距 \(\beta_{0j}\) 和随机斜率 \(\beta_{1j}\),指标 \(j\) 表示分组的编号,也叫变截距和变斜率模型
\[ \begin{aligned} \mathrm{Reaction}_{ij} &= \beta_{0j} + \beta_{1j} \cdot \mathrm{Days}_{ij} + \epsilon_{ij} \\ \beta_{0j} &= \gamma_{00} + U_{0j} \\ \beta_{1j} &= \gamma_{10} + U_{1j} \\ \begin{pmatrix} U_{0j} \\ U_{1j} \end{pmatrix} &\sim \mathcal{N} \begin{bmatrix} \begin{pmatrix} 0 \\ 0 \end{pmatrix} , \begin{pmatrix} \tau^2_{00} & \tau_{01} \\ \tau_{01} & \tau^2_{10} \end{pmatrix} \end{bmatrix} \\ \epsilon_{ij} &\sim \mathcal{N}(0, \sigma^2) \\ i = 0,1,\cdots,9 &\quad j = 308,309,\cdots, 372. \end{aligned} \]
下面用 nlme 包 (Pinheiro 和 Bates 2000) 拟合模型。
library(nlme)
sleep_nlme <- lme(Reaction ~ Days, random = ~ Days | Subject, data = sleepstudy)
summary(sleep_nlme)#> Linear mixed-effects model fit by REML
#> Data: sleepstudy
#> AIC BIC logLik
#> 1755.628 1774.719 -871.8141
#>
#> Random effects:
#> Formula: ~Days | Subject
#> Structure: General positive-definite, Log-Cholesky parametrization
#> StdDev Corr
#> (Intercept) 24.740241 (Intr)
#> Days 5.922103 0.066
#> Residual 25.591843
#>
#> Fixed effects: Reaction ~ Days
#> Value Std.Error DF t-value p-value
#> (Intercept) 251.40510 6.824516 161 36.83853 0
#> Days 10.46729 1.545783 161 6.77151 0
#> Correlation:
#> (Intr)
#> Days -0.138
#>
#> Standardized Within-Group Residuals:
#> Min Q1 Med Q3 Max
#> -3.95355735 -0.46339976 0.02311783 0.46339621 5.17925089
#>
#> Number of Observations: 180
#> Number of Groups: 18随机效应(Random effects)部分:
#> (Intercept) Days
#> 308 2.258754 9.1989366
#> 309 -40.398490 -8.6197167
#> 310 -38.960098 -5.4489048
#> 330 23.690228 -4.8142826
#> 331 22.259981 -3.0698548
#> 332 9.039458 -0.2721585固定效应(Fixed effects)部分:
ggeffects 包的函数 ggpredict() 和 ggeffect() 可以用来绘制混合效应模型的边际效应( Marginal Effects),ggPMX 包 可以用来绘制混合效应模型的诊断图。下 图 36.3 展示关于变量 Days 的边际效应图。
36.1.2 MASS
sleep_mass <- MASS::glmmPQL(Reaction ~ Days,
random = ~ Days | Subject, verbose = FALSE,
data = sleepstudy, family = gaussian
)
summary(sleep_mass)#> Linear mixed-effects model fit by maximum likelihood
#> Data: sleepstudy
#> AIC BIC logLik
#> NA NA NA
#>
#> Random effects:
#> Formula: ~Days | Subject
#> Structure: General positive-definite, Log-Cholesky parametrization
#> StdDev Corr
#> (Intercept) 23.780376 (Intr)
#> Days 5.716807 0.081
#> Residual 25.591842
#>
#> Variance function:
#> Structure: fixed weights
#> Formula: ~invwt
#> Fixed effects: Reaction ~ Days
#> Value Std.Error DF t-value p-value
#> (Intercept) 251.40510 6.669396 161 37.69533 0
#> Days 10.46729 1.510647 161 6.92901 0
#> Correlation:
#> (Intr)
#> Days -0.138
#>
#> Standardized Within-Group Residuals:
#> Min Q1 Med Q3 Max
#> -3.94156355 -0.46559311 0.02894656 0.46361051 5.17933587
#>
#> Number of Observations: 180
#> Number of Groups: 1836.1.3 lme4
#> Linear mixed model fit by REML ['lmerMod']
#> Formula: Reaction ~ Days + (Days | Subject)
#> Data: sleepstudy
#>
#> REML criterion at convergence: 1743.6
#>
#> Scaled residuals:
#> Min 1Q Median 3Q Max
#> -3.9536 -0.4634 0.0231 0.4634 5.1793
#>
#> Random effects:
#> Groups Name Variance Std.Dev. Corr
#> Subject (Intercept) 612.10 24.741
#> Days 35.07 5.922 0.07
#> Residual 654.94 25.592
#> Number of obs: 180, groups: Subject, 18
#>
#> Fixed effects:
#> Estimate Std. Error t value
#> (Intercept) 251.405 6.825 36.838
#> Days 10.467 1.546 6.771
#>
#> Correlation of Fixed Effects:
#> (Intr)
#> Days -0.13836.1.4 blme
sleep_blme <- blme::blmer(
Reaction ~ Days + (Days | Subject), data = sleepstudy,
control = lme4::lmerControl(check.conv.grad = "ignore"),
cov.prior = NULL)
summary(sleep_blme)#> Prior dev : 0
#>
#> Linear mixed model fit by REML ['blmerMod']
#> Formula: Reaction ~ Days + (Days | Subject)
#> Data: sleepstudy
#> Control: lme4::lmerControl(check.conv.grad = "ignore")
#>
#> REML criterion at convergence: 1743.6
#>
#> Scaled residuals:
#> Min 1Q Median 3Q Max
#> -3.9536 -0.4634 0.0231 0.4634 5.1793
#>
#> Random effects:
#> Groups Name Variance Std.Dev. Corr
#> Subject (Intercept) 612.10 24.741
#> Days 35.07 5.922 0.07
#> Residual 654.94 25.592
#> Number of obs: 180, groups: Subject, 18
#>
#> Fixed effects:
#> Estimate Std. Error t value
#> (Intercept) 251.405 6.825 36.838
#> Days 10.467 1.546 6.771
#>
#> Correlation of Fixed Effects:
#> (Intr)
#> Days -0.13836.1.5 brms
Family: gaussian
Links: mu = identity; sigma = identity
Formula: Reaction ~ Days + (Days | Subject)
Data: sleepstudy (Number of observations: 180)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Group-Level Effects:
~Subject (Number of levels: 18)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sd(Intercept) 27.03 6.60 15.88 42.13 1.00 1728 2469
sd(Days) 6.61 1.50 4.18 9.97 1.00 1517 2010
cor(Intercept,Days) 0.08 0.29 -0.46 0.65 1.00 991 1521
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept 251.26 7.42 236.27 266.12 1.00 1982 2687
Days 10.36 1.77 6.85 13.85 1.00 1415 1982
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 25.88 1.54 22.99 29.06 1.00 3204 2869
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).36.1.6 MCMCglmm
将数据集 sleepstudy 中的 Reaction 除以 1000,目的是数值稳定性,减小迭代序列的相关性。
## 尺度变换
sleepstudy$Reaction <- sleepstudy$Reaction / 1000
## 变截距、变斜率模型
prior1 <- list(
R = list(V = 1, nu = 0.002),
G = list(G1 = list(V = diag(2), nu = 0.002))
)
set.seed(20232023)
sleep_mcmcglmm <- MCMCglmm::MCMCglmm(
Reaction ~ Days, random = ~ us(1 + Days):Subject, prior = prior1,
data = sleepstudy, family = "gaussian", verbose = FALSE
)
summary(sleep_mcmcglmm)#>
#> Iterations = 3001:12991
#> Thinning interval = 10
#> Sample size = 1000
#>
#> DIC: -774.797
#>
#> G-structure: ~us(1 + Days):Subject
#>
#> post.mean l-95% CI u-95% CI eff.samp
#> (Intercept):(Intercept).Subject 9.848e-04 3.046e-04 0.0019260 1000.0
#> Days:(Intercept).Subject -2.678e-05 -2.685e-04 0.0002223 1091.0
#> (Intercept):Days.Subject -2.678e-05 -2.685e-04 0.0002223 1091.0
#> Days:Days.Subject 1.971e-04 8.733e-05 0.0003453 878.8
#>
#> R-structure: ~units
#>
#> post.mean l-95% CI u-95% CI eff.samp
#> units 0.0006742 0.0005304 0.0008345 909.7
#>
#> Location effects: Reaction ~ Days
#>
#> post.mean l-95% CI u-95% CI eff.samp pMCMC
#> (Intercept) 0.25167 0.23608 0.26961 1000 <0.001 ***
#> Days 0.01055 0.00334 0.01650 1000 0.008 **
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1固定效应 Location effects 截距 (Intercept) 为 0.25167,斜率 Days 为 0.01055 。
随机效应 R-structure 方差 0.0006742,则标准差为 0.02596536。
36.1.7 INLA
先考虑变截距模型
library(INLA)
inla.setOption(short.summary = TRUE)
# 前面已经做了尺度变换
# sleepstudy$Reaction <- sleepstudy$Reaction / 1000
# 变截距
sleep_inla1 <- inla(Reaction ~ Days + f(Subject, model = "iid", n = 18),
family = "gaussian", data = sleepstudy)
# 输出结果
summary(sleep_inla1)#> Fixed effects:
#> mean sd 0.025quant 0.5quant 0.975quant mode kld
#> (Intercept) 0.251 0.010 0.232 0.251 0.270 0.251 0
#> Days 0.010 0.001 0.009 0.010 0.012 0.010 0
#>
#> Model hyperparameters:
#> mean sd 0.025quant 0.5quant
#> Precision for the Gaussian observations 1054.07 116.99 840.14 1048.46
#> Precision for Subject 843.97 300.88 391.25 798.36
#> 0.975quant mode
#> Precision for the Gaussian observations 1300.13 1038.99
#> Precision for Subject 1559.54 714.37
#>
#> is computed再考虑变截距和变斜率模型
# https://inla.r-inla-download.org/r-inla.org/doc/latent/iid.pdf
# 二维高斯随机效应的先验为 Wishart prior
sleepstudy$Subject <- as.integer(sleepstudy$Subject)
sleepstudy$slopeid <- 18 + sleepstudy$Subject
# 变截距、变斜率
sleep_inla2 <- inla(
Reaction ~ 1 + Days + f(Subject, model = "iid2d", n = 2 * 18) + f(slopeid, Days, copy = "Subject"),
data = sleepstudy, family = "gaussian"
)
# 输出结果
summary(sleep_inla2)#> Fixed effects:
#> mean sd 0.025quant 0.5quant 0.975quant mode kld
#> (Intercept) 0.251 0.055 0.142 0.251 0.360 0.251 0
#> Days 0.010 0.054 -0.097 0.010 0.118 0.010 0
#>
#> Model hyperparameters:
#> mean sd 0.025quant 0.5quant
#> Precision for the Gaussian observations 1549.518 181.248 1218.040 1540.851
#> Precision for Subject (component 1) 20.871 6.507 10.626 20.024
#> Precision for Subject (component 2) 21.224 6.611 10.804 20.367
#> Rho1:2 for Subject -0.001 0.213 -0.414 -0.002
#> 0.975quant mode
#> Precision for the Gaussian observations 1930.556 1527.278
#> Precision for Subject (component 1) 35.971 18.479
#> Precision for Subject (component 2) 36.555 18.806
#> Rho1:2 for Subject 0.413 -0.003
#>
#> is computed36.2 广义线性混合效应模型
当响应变量分布不再是高斯分布,线性混合效应模型就扩展到广义线性混合效应模型。有一些 R 包可以拟合此类模型,MASS 包的函数 glmmPQL() ,mgcv 包的函数 gam(),lme4 包的函数 glmer() ,GLMMadaptive 包的函数 mixed_model() ,brms 包的函数 brm() 等。
| 响应变量分布 | MASS | mgcv | lme4 | GLMMadaptive | brms |
|---|---|---|---|---|---|
| 伯努利分布 | 支持 | 支持 | 支持 | 支持 | 支持 |
| 二项分布 | 支持 | 支持 | 支持 | 支持 | 支持 |
| 泊松分布 | 支持 | 支持 | 支持 | 支持 | 支持 |
| 负二项分布 | 不支持 | 支持 | 支持 | 支持 | 支持 |
| 伽马分布 | 支持 | 支持 | 支持 | 支持 | 支持 |
函数 glmmPQL() 支持的分布族见函数 glm() 的参数 family ,lme4 包的函数 glmer.nb() 和 GLMMadaptive 包的函数 negative.binomial() 都可用于拟合响应变量服从负二项分布的情况。除了这些常规的分布,GLMMadaptive 和 brms 包还支持许多常见的分布,比如零膨胀的泊松分布、二项分布等,还可以自定义分布。
- 伯努利分布
family = binomial(link = "logit") - 二项分布
family = binomial(link = "logit") - 泊松分布
family = poisson(link = "log") - 负二项分布
lme4::glmer.nb()或GLMMadaptive::negative.binomial() - 伽马分布
family = Gamma(link = "inverse")
GLMMadaptive 包 (Rizopoulos 2023) 的主要函数 mixed_model() 是用来拟合广义线性混合效应模型的。下面以牛传染性胸膜肺炎(Contagious bovine pleuropneumonia,简称 CBPP)数据 cbpp 介绍函数 mixed_model() 的用法,该数据集来自 lme4 包。
#> 'data.frame': 56 obs. of 4 variables:
#> $ herd : Factor w/ 15 levels "1","2","3","4",..: 1 1 1 1 2 2 2 3 3 3 ...
#> $ incidence: num 2 3 4 0 3 1 1 8 2 0 ...
#> $ size : num 14 12 9 5 22 18 21 22 16 16 ...
#> $ period : Factor w/ 4 levels "1","2","3","4": 1 2 3 4 1 2 3 1 2 3 ...herd 牛群编号,period 时间段,incidence 感染的数量,size 牛群大小。疾病在种群内扩散
ggplot(data = cbpp, aes(x = herd, y = period)) +
geom_tile(aes(fill = incidence / size)) +
scale_fill_viridis_c(label = scales::percent_format(),
option = "C", name = "") +
theme_minimal()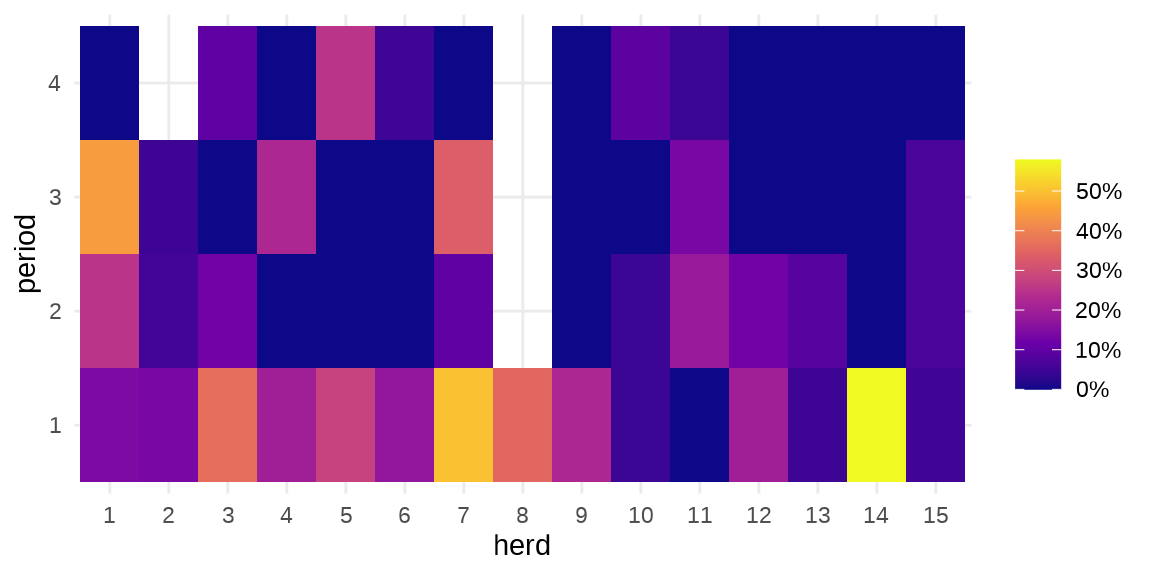
36.2.1 MASS
cbpp_mass <- MASS::glmmPQL(
cbind(incidence, size - incidence) ~ period,
random = ~ 1 | herd, verbose = FALSE,
data = cbpp, family = binomial("logit")
)
summary(cbpp_mass)#> Linear mixed-effects model fit by maximum likelihood
#> Data: cbpp
#> AIC BIC logLik
#> NA NA NA
#>
#> Random effects:
#> Formula: ~1 | herd
#> (Intercept) Residual
#> StdDev: 0.5563535 1.184527
#>
#> Variance function:
#> Structure: fixed weights
#> Formula: ~invwt
#> Fixed effects: cbind(incidence, size - incidence) ~ period
#> Value Std.Error DF t-value p-value
#> (Intercept) -1.327364 0.2390194 38 -5.553372 0.0000
#> period2 -1.016126 0.3684079 38 -2.758156 0.0089
#> period3 -1.149984 0.3937029 38 -2.920944 0.0058
#> period4 -1.605217 0.5178388 38 -3.099839 0.0036
#> Correlation:
#> (Intr) perid2 perid3
#> period2 -0.399
#> period3 -0.373 0.260
#> period4 -0.282 0.196 0.182
#>
#> Standardized Within-Group Residuals:
#> Min Q1 Med Q3 Max
#> -2.0591168 -0.6493095 -0.2747620 0.5170492 2.6187632
#>
#> Number of Observations: 56
#> Number of Groups: 1536.2.2 GLMMadaptive
library(GLMMadaptive)
cbpp_glmmadaptive <- mixed_model(
fixed = cbind(incidence, size - incidence) ~ period,
random = ~ 1 | herd, data = cbpp, family = binomial(link = "logit")
)
summary(cbpp_glmmadaptive)#>
#> Call:
#> mixed_model(fixed = cbind(incidence, size - incidence) ~ period,
#> random = ~1 | herd, data = cbpp, family = binomial(link = "logit"))
#>
#> Data Descriptives:
#> Number of Observations: 56
#> Number of Groups: 15
#>
#> Model:
#> family: binomial
#> link: logit
#>
#> Fit statistics:
#> log.Lik AIC BIC
#> -91.98337 193.9667 197.507
#>
#> Random effects covariance matrix:
#> StdDev
#> (Intercept) 0.6475934
#>
#> Fixed effects:
#> Estimate Std.Err z-value p-value
#> (Intercept) -1.3995 0.2335 -5.9923 < 1e-04
#> period2 -0.9914 0.3068 -3.2316 0.00123091
#> period3 -1.1278 0.3268 -3.4513 0.00055793
#> period4 -1.5795 0.4276 -3.6937 0.00022101
#>
#> Integration:
#> method: adaptive Gauss-Hermite quadrature rule
#> quadrature points: 11
#>
#> Optimization:
#> method: EM
#> converged: TRUE36.2.3 glmmTMB
cbpp_glmmtmb <- glmmTMB::glmmTMB(
cbind(incidence, size - incidence) ~ period + (1 | herd),
data = cbpp, family = binomial, REML = TRUE
)
summary(cbpp_glmmtmb)#> Family: binomial ( logit )
#> Formula: cbind(incidence, size - incidence) ~ period + (1 | herd)
#> Data: cbpp
#>
#> AIC BIC logLik deviance df.resid
#> 196.4 206.5 -93.2 186.4 55
#>
#> Random effects:
#>
#> Conditional model:
#> Groups Name Variance Std.Dev.
#> herd (Intercept) 0.4649 0.6819
#> Number of obs: 56, groups: herd, 15
#>
#> Conditional model:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -1.3670 0.2376 -5.752 8.79e-09 ***
#> period2 -0.9693 0.3055 -3.173 0.001509 **
#> period3 -1.1045 0.3255 -3.393 0.000691 ***
#> period4 -1.5519 0.4265 -3.639 0.000274 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 136.2.4 lme4
cbpp_lme4 <- lme4::glmer(
cbind(incidence, size - incidence) ~ period + (1 | herd),
family = binomial("logit"), data = cbpp
)
summary(cbpp_lme4)#> Generalized linear mixed model fit by maximum likelihood (Laplace
#> Approximation) [glmerMod]
#> Family: binomial ( logit )
#> Formula: cbind(incidence, size - incidence) ~ period + (1 | herd)
#> Data: cbpp
#>
#> AIC BIC logLik deviance df.resid
#> 194.1 204.2 -92.0 184.1 51
#>
#> Scaled residuals:
#> Min 1Q Median 3Q Max
#> -2.3816 -0.7889 -0.2026 0.5142 2.8791
#>
#> Random effects:
#> Groups Name Variance Std.Dev.
#> herd (Intercept) 0.4123 0.6421
#> Number of obs: 56, groups: herd, 15
#>
#> Fixed effects:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -1.3983 0.2312 -6.048 1.47e-09 ***
#> period2 -0.9919 0.3032 -3.272 0.001068 **
#> period3 -1.1282 0.3228 -3.495 0.000474 ***
#> period4 -1.5797 0.4220 -3.743 0.000182 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Correlation of Fixed Effects:
#> (Intr) perid2 perid3
#> period2 -0.363
#> period3 -0.340 0.280
#> period4 -0.260 0.213 0.19836.2.5 mgcv
或使用 mgcv 包,可以得到近似的结果。随机效应部分可以看作可加的惩罚项
library(mgcv)
cbpp_mgcv <- gam(
cbind(incidence, size - incidence) ~ period + s(herd, bs = "re"),
data = cbpp, family = binomial(link = "logit"), method = "REML"
)
summary(cbpp_mgcv)#>
#> Family: binomial
#> Link function: logit
#>
#> Formula:
#> cbind(incidence, size - incidence) ~ period + s(herd, bs = "re")
#>
#> Parametric coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -1.3670 0.2358 -5.799 6.69e-09 ***
#> period2 -0.9693 0.3040 -3.189 0.001428 **
#> period3 -1.1045 0.3241 -3.407 0.000656 ***
#> period4 -1.5519 0.4251 -3.651 0.000261 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Approximate significance of smooth terms:
#> edf Ref.df Chi.sq p-value
#> s(herd) 9.66 14 32.03 3.21e-05 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> R-sq.(adj) = 0.515 Deviance explained = 53%
#> -REML = 93.199 Scale est. = 1 n = 56下面给出随机效应的标准差的估计及其上下限,和前面 GLMMadaptive 包和 lme4 包给出的结果也是接近的。
36.2.6 blme
cbpp_blme <- blme::bglmer(
cbind(incidence, size - incidence) ~ period + (1 | herd),
family = binomial("logit"), data = cbpp
)
summary(cbpp_blme)#> Cov prior : herd ~ wishart(df = 3.5, scale = Inf, posterior.scale = cov, common.scale = TRUE)
#> Prior dev : 0.9901
#>
#> Generalized linear mixed model fit by maximum likelihood (Laplace
#> Approximation) [bglmerMod]
#> Family: binomial ( logit )
#> Formula: cbind(incidence, size - incidence) ~ period + (1 | herd)
#> Data: cbpp
#>
#> AIC BIC logLik deviance df.resid
#> 194.2 204.3 -92.1 184.2 51
#>
#> Scaled residuals:
#> Min 1Q Median 3Q Max
#> -2.3670 -0.8121 -0.1704 0.4971 2.7969
#>
#> Random effects:
#> Groups Name Variance Std.Dev.
#> herd (Intercept) 0.5168 0.7189
#> Number of obs: 56, groups: herd, 15
#>
#> Fixed effects:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -1.4142 0.2466 -5.736 9.7e-09 ***
#> period2 -0.9803 0.3037 -3.227 0.001249 **
#> period3 -1.1171 0.3233 -3.455 0.000549 ***
#> period4 -1.5667 0.4222 -3.710 0.000207 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Correlation of Fixed Effects:
#> (Intr) perid2 perid3
#> period2 -0.342
#> period3 -0.320 0.282
#> period4 -0.246 0.215 0.19936.2.7 brms
表示二项分布,公式语法与前面的 lme4 等包不同。
Family: binomial
Links: mu = logit
Formula: incidence | trials(size) ~ period + (1 | herd)
Data: cbpp (Number of observations: 56)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Group-Level Effects:
~herd (Number of levels: 15)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sd(Intercept) 0.76 0.22 0.39 1.29 1.00 1483 1962
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept -1.40 0.26 -1.92 -0.88 1.00 2440 2542
period2 -1.00 0.31 -1.63 -0.41 1.00 5242 2603
period3 -1.14 0.34 -1.83 -0.50 1.00 4938 3481
period4 -1.61 0.44 -2.49 -0.81 1.00 4697 2966
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).36.2.8 MCMCglmm
set.seed(20232023)
cbpp_mcmcglmm <- MCMCglmm::MCMCglmm(
cbind(incidence, size - incidence) ~ period, random = ~herd,
data = cbpp, family = "multinomial2", verbose = FALSE
)
summary(cbpp_mcmcglmm)#>
#> Iterations = 3001:12991
#> Thinning interval = 10
#> Sample size = 1000
#>
#> DIC: 538.4141
#>
#> G-structure: ~herd
#>
#> post.mean l-95% CI u-95% CI eff.samp
#> herd 0.0244 1.256e-16 0.1347 186.5
#>
#> R-structure: ~units
#>
#> post.mean l-95% CI u-95% CI eff.samp
#> units 1.098 0.2471 2.158 273.9
#>
#> Location effects: cbind(incidence, size - incidence) ~ period
#>
#> post.mean l-95% CI u-95% CI eff.samp pMCMC
#> (Intercept) -1.5314 -2.1484 -0.8507 1000.0 <0.001 ***
#> period2 -1.2596 -2.2129 -0.1495 854.7 0.006 **
#> period3 -1.3827 -2.3979 -0.2851 691.0 0.012 *
#> period4 -1.9612 -3.3031 -0.7745 572.6 <0.001 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1对于服从非高斯分布的响应变量,MCMCglmm 总是假定存在过度离散的情况,即存在一个与分类变量无关的随机变量,或者说存在一个残差服从正态分布的随机变量(效应),可以看作测量误差,这种假定对真实数据建模是有意义的,所以,与以上 MCMCglmm 代码等价的 lme4 包模型代码如下:
cbpp$id <- as.factor(1:dim(cbpp)[1])
cbpp_lme4 <- lme4::glmer(
cbind(incidence, size - incidence) ~ period + (1 | herd) + (1 | id),
family = binomial, data = cbpp
)
summary(cbpp_lme4)#> Generalized linear mixed model fit by maximum likelihood (Laplace
#> Approximation) [glmerMod]
#> Family: binomial ( logit )
#> Formula: cbind(incidence, size - incidence) ~ period + (1 | herd) + (1 |
#> id)
#> Data: cbpp
#>
#> AIC BIC logLik deviance df.resid
#> 186.6 198.8 -87.3 174.6 50
#>
#> Scaled residuals:
#> Min 1Q Median 3Q Max
#> -1.2866 -0.5989 -0.1181 0.3575 1.6216
#>
#> Random effects:
#> Groups Name Variance Std.Dev.
#> id (Intercept) 0.79400 0.8911
#> herd (Intercept) 0.03384 0.1840
#> Number of obs: 56, groups: id, 56; herd, 15
#>
#> Fixed effects:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -1.5003 0.2967 -5.056 4.27e-07 ***
#> period2 -1.2265 0.4803 -2.554 0.01066 *
#> period3 -1.3288 0.4939 -2.690 0.00713 **
#> period4 -1.8662 0.5936 -3.144 0.00167 **
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Correlation of Fixed Effects:
#> (Intr) perid2 perid3
#> period2 -0.559
#> period3 -0.537 0.373
#> period4 -0.441 0.327 0.314贝叶斯的结果与频率派的结果相近,但还是有明显差异。
36.2.9 INLA
表示二项分布,公式语法与前面的 brms 包和 lme4 等包都不同。
cbpp_inla <- inla(
formula = incidence ~ period + f(herd, model = "iid", n = 15),
Ntrials = size, family = "binomial", data = cbpp
)
summary(cbpp_inla)#> Fixed effects:
#> mean sd 0.025quant 0.5quant 0.975quant mode kld
#> (Intercept) -1.382 0.224 -1.845 -1.375 -0.957 -1.375 0
#> period2 -1.032 0.304 -1.628 -1.032 -0.434 -1.032 0
#> period3 -1.174 0.324 -1.809 -1.174 -0.537 -1.174 0
#> period4 -1.662 0.425 -2.495 -1.662 -0.827 -1.662 0
#>
#> Model hyperparameters:
#> mean sd 0.025quant 0.5quant 0.975quant mode
#> Precision for herd 5.10 6.39 1.02 3.29 18.55 2.17
#>
#> is computed36.3 非线性混合效应模型
Loblolly 数据集来自 R 内置的 datasets 包
36.3.1 nlme
非线性回归
nfm1 <- nls(height ~ SSasymp(age, Asym, R0, lrc),
data = Loblolly, subset = Seed == 329)
summary(nfm1)#>
#> Formula: height ~ SSasymp(age, Asym, R0, lrc)
#>
#> Parameters:
#> Estimate Std. Error t value Pr(>|t|)
#> Asym 94.1282 8.4030 11.202 0.001525 **
#> R0 -8.2508 1.2261 -6.729 0.006700 **
#> lrc -3.2176 0.1386 -23.218 0.000175 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 0.7493 on 3 degrees of freedom
#>
#> Number of iterations to convergence: 0
#> Achieved convergence tolerance: 3.919e-07非线性函数 SSasymp() 的内容如下
\[ \mathrm{Asym}+(\mathrm{R0}-\mathrm{Asym})\times\exp\big(-\exp(\mathrm{lrc})\times\mathrm{input}\big) \]
其中,\(\mathrm{Asym}\) 、\(\mathrm{R0}\) 、\(\mathrm{lrc}\) 是参数,\(\mathrm{input}\) 是输入值。
示例来自 nlme 包的函数 nlme() 帮助文档
nfm2 <- nlme(height ~ SSasymp(age, Asym, R0, lrc),
data = Loblolly,
fixed = Asym + R0 + lrc ~ 1,
random = Asym ~ 1,
start = c(Asym = 103, R0 = -8.5, lrc = -3.3)
)
summary(nfm2)#> Nonlinear mixed-effects model fit by maximum likelihood
#> Model: height ~ SSasymp(age, Asym, R0, lrc)
#> Data: Loblolly
#> AIC BIC logLik
#> 239.4856 251.6397 -114.7428
#>
#> Random effects:
#> Formula: Asym ~ 1 | Seed
#> Asym Residual
#> StdDev: 3.650642 0.7188625
#>
#> Fixed effects: Asym + R0 + lrc ~ 1
#> Value Std.Error DF t-value p-value
#> Asym 101.44960 2.4616951 68 41.21128 0
#> R0 -8.62733 0.3179505 68 -27.13420 0
#> lrc -3.23375 0.0342702 68 -94.36052 0
#> Correlation:
#> Asym R0
#> R0 0.704
#> lrc -0.908 -0.827
#>
#> Standardized Within-Group Residuals:
#> Min Q1 Med Q3 Max
#> -2.23601930 -0.62380854 0.05917466 0.65727206 1.95794425
#>
#> Number of Observations: 84
#> Number of Groups: 14#> Nonlinear mixed-effects model fit by maximum likelihood
#> Model: height ~ SSasymp(age, Asym, R0, lrc)
#> Data: Loblolly
#> AIC BIC logLik
#> 238.9662 253.5511 -113.4831
#>
#> Random effects:
#> Formula: list(Asym ~ 1, lrc ~ 1)
#> Level: Seed
#> Structure: Diagonal
#> Asym lrc Residual
#> StdDev: 2.806185 0.03449969 0.6920003
#>
#> Fixed effects: Asym + R0 + lrc ~ 1
#> Value Std.Error DF t-value p-value
#> Asym 101.85205 2.3239828 68 43.82651 0
#> R0 -8.59039 0.3058441 68 -28.08747 0
#> lrc -3.24011 0.0345017 68 -93.91167 0
#> Correlation:
#> Asym R0
#> R0 0.727
#> lrc -0.902 -0.796
#>
#> Standardized Within-Group Residuals:
#> Min Q1 Med Q3 Max
#> -2.06072906 -0.69785679 0.08721706 0.73687722 1.79015782
#>
#> Number of Observations: 84
#> Number of Groups: 1436.3.2 lme4
lme4 的公式语法是与 nlme 包不同的。
lob_lme4 <- lme4::nlmer(
height ~ SSasymp(age, Asym, R0, lrc) ~ (Asym + R0 + lrc) + (Asym | Seed),
data = Loblolly,
start = c(Asym = 103, R0 = -8.5, lrc = -3.3)
)
summary(lob_lme4)#> Nonlinear mixed model fit by maximum likelihood ['nlmerMod']
#> Formula: height ~ SSasymp(age, Asym, R0, lrc) ~ (Asym + R0 + lrc) + (Asym |
#> Seed)
#> Data: Loblolly
#>
#> AIC BIC logLik deviance df.resid
#> 239.4 251.5 -114.7 229.4 79
#>
#> Scaled residuals:
#> Min 1Q Median 3Q Max
#> -2.24149 -0.62546 0.08326 0.67711 1.91351
#>
#> Random effects:
#> Groups Name Variance Std.Dev.
#> Seed Asym 13.5121 3.6759
#> Residual 0.5161 0.7184
#> Number of obs: 84, groups: Seed, 14
#>
#> Fixed effects:
#> Estimate Std. Error t value
#> Asym 102.119832 0.012378 8249.9
#> R0 -8.549401 0.012354 -692.0
#> lrc -3.243973 0.008208 -395.2
#>
#> Correlation of Fixed Effects:
#> Asym R0
#> R0 0.000
#> lrc -0.008 -0.03436.3.3 brms
根据数据的情况,设定参数的先验分布
根据模型表达式编码
Family: gaussian
Links: mu = identity; sigma = identity
Formula: height ~ Asym + (R0 - Asym) * exp(-exp(lrc) * age)
R0 ~ 1
lrc ~ 1
Asym ~ 1 + (1 | Seed)
Data: Loblolly (Number of observations: 84)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Group-Level Effects:
~Seed (Number of levels: 14)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sd(Asym_Intercept) 3.90 1.09 2.24 6.51 1.00 1033 1647
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
R0_Intercept -8.53 0.43 -9.37 -7.68 1.00 2236 1434
lrc_Intercept -3.23 0.02 -3.27 -3.20 1.00 981 1546
Asym_Intercept 101.00 0.10 100.80 101.20 1.00 4443 2907
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 1.68 0.25 1.20 2.17 1.00 1910 2258
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).36.4 总结
本章介绍函数 MASS::glmmPQL()、 nlme::lme()、lme4::lmer() 和 brms::brm() 的用法,以及它们求解线性混合效应模型的区别和联系。在贝叶斯估计方法中,brms 包和 INLA 包都支持非常丰富的模型种类,前者是贝叶斯精确推断,后者是贝叶斯近似推断,brms 基于概率编程语言 Stan 框架打包了许多模型的 Stan 实现,INLA 基于求解随机偏微分方程的有限元方法和拉普拉斯近似技巧,将各类常见统计模型统一起来,计算速度快,计算结果准确。
- 函数
nlme::lme()极大似然估计和限制极大似然估计 - 函数
MASS::glmmPQL()惩罚拟似然估计,MASS 是依赖 nlme 包, nlme 不支持模型中添加漂移项,所以函数glmmPQL()也不支持添加漂移项。 - 函数
lme4::lmer()拉普拉斯近似。关于随机效应的高维积分 - 函数
brms::brm()汉密尔顿蒙特卡罗抽样。HMC 方法结合自适应步长的采样器 NUTS 来抽样。 - 函数
INLA::inla()集成嵌套拉普拉斯近似。
| 模型 | nlme | MASS | lme4 | GLMMadaptive | brms |
|---|---|---|---|---|---|
| 线性混合效应模型 | lme() |
glmmPQL() |
lmer() |
不支持 | brm() |
| 广义线性混合效应模型 | 不支持 | glmmPQL() |
glmer() |
mixed_model() |
brm() |
| 非线性混合效应模型 | nlme() |
不支持 | nlmer() |
不支持 | brm() |
通过对频率派和贝叶斯派方法的比较,发现一些有意思的结果。与 Stan 不同,INLA 包做近似贝叶斯推断,计算效率很高。
INLA 软件能处理上千个高斯随机效应,但最多只能处理 15 个超参数,因为 INLA 使用 CCD 处理超参数。如果使用 MCMC 处理超参数,就有可能处理更多的超参数,Daniel Simpson 等把 Laplace approximation 带入 Stan,这样就可以处理上千个超参数。 更多理论内容见 2009 年 INLA 诞生的论文和《Advanced Spatial Modeling with Stochastic Partial Differential Equations Using R and INLA》中第一章的估计方法 CCD。
36.5 习题
基于奥克兰火山地形数据集 volcano ,随机拆分成训练数据和测试数据,训练数据可以看作采样点的观测数据,建立高斯过程回归模型,比较测试数据与未采样的位置上的预测数据,在计算速度、准确度、易用性等方面总结 Stan 和 INLA 的特点。
-
基于
PlantGrowth数据集,比较将group变量视为随机变量与随机效应的异同? -
从广义线性混合效应模型生成模拟数据,用至少 6 个不同的 R 包估计模型参数,比较和归纳不同估计方法和实现算法的效果。举例:带漂移项的泊松型广义线性混合效应模型。\(y_{ij}\) 表示响应变量,\(\bm{u}\) 表示随机效应,\(o_{ij}\) 表示漂移项。
\[ \begin{aligned} y_{ij}|\bm{u} &\sim \mathrm{Poisson}(o_{ij}\lambda_{ij}) \\ \log(\lambda_{ij}) &= \beta_{ij}x_{ij} + u_{j} \\ u_j &\sim \mathcal{N}(0, \sigma^2) \\ i = 1,2,\ldots, n &\quad j = 1,2,\ldots,q \end{aligned} \]
代码
set.seed(2023) Ngroups <- 25 # 一个随机效应分 25 个组 NperGroup <- 100 # 每个组 100 个观察值 # 样本量 N <- Ngroups * NperGroup # 截距和两个协变量的系数 beta <- c(0.5, 0.3, 0.2) # 两个协变量 X <- MASS::mvrnorm(N, mu = rep(0, 2), Sigma = matrix(c(1, 0.8, 0.8, 1), 2)) # 漂移项 o <- rep(c(2, 4), each = N / 2) # 分 25 个组 每个组 100 个观察值 g <- factor(rep(1:Ngroups, each = NperGroup)) u <- rnorm(Ngroups, sd = .5) # 随机效应的标准差 0.5 # 泊松分布的期望 lambda <- o * exp(cbind(1, X) %*% beta + u[g]) # 响应变量的值 y <- rpois(N, lambda = lambda) # 模拟的数据集 sim_data <- data.frame(y, X, o, g) colnames(sim_data) <- c("y", "x1", "x2", "o", "g") # 模型拟合 fit_lme4 <- lme4::glmer(y ~ x1 + x2 + (1 | g), data = sim_data, offset = log(o), family = poisson(link = "log") ) summary(fit_lme4) # 对随机效应 adaptive Gauss-Hermite quadrature 积分 library(GLMMadaptive) fit_glmmadaptive <- mixed_model( fixed = y ~ x1 + x2 + offset(log(o)), random = ~ 1 | g, data = sim_data, family = poisson(link = "log") ) summary(fit_glmmadaptive) ## hglm 包 Hierarchical Generalized Linear Models # extended quasi likelihood (EQL) method fit_hglm <- hglm::hglm( fixed = y ~ x1 + x2, random = ~ 1 | g, family = poisson(link = "log"), offset = log(o), data = sim_data ) summary(fit_hglm) # 广义估计方程 fit_gee <- gee::gee(y ~ x1 + x2 + offset(log(o)), id = g, data = sim_data, family = poisson(link = "log"), corstr = "exchangeable" ) # 输出 fit_gee # [glmmML](https://CRAN.R-project.org/package=glmmML) # Maximum Likelihood and numerical integration via Gauss-Hermite quadrature. fit_glmmml <- glmmML::glmmML( formula = y ~ x1 + x2, family = poisson, data = sim_data, offset = log(o), cluster = g ) summary(fit_glmmml) # Generalized Estimating Equation # [geepack](https://cran.r-project.org/package=geepack) GEE fit_geepack <- geepack::geeglm( formula = y ~ x1 + x2, family = poisson(link = "log"), id = g, offset = log(o), data = sim_data, corstr = "exchangeable", scale.fix = FALSE ) summary(fit_geepack) # [glmm](https://github.com/knudson1/glmm) # Monte Carlo Likelihood Approximation 近似对随机效应的积分 # 对迭代时间没有给出预估,一旦执行,不知道什么时候会跑完 set.seed(2023) # 设置双核并行迭代 clust <- parallel::makeCluster(2) fit_glmm <- glmm::glmm(y ~ x1 + x2 + offset(log(o)), random = list(~ 1 + g), # 随机效应 varcomps.names = "G", # 给随机效应取个名字 data = sim_data, family.glmm = glmm::poisson.glmm, # 泊松型 m = 10^4, debug = TRUE, cluster = clust ) parallel::stopCluster(clust) summary(fit_glmm) # 如何设置先验分布 fit_brms <- brms::brm( y ~ x1 + x2 + (1 | g) + offset(log(o)), data = sim_data, family = poisson(link = "log"), # silent = 2, # 关闭消息 refresh = 0, # 不显示迭代进度 seed = 20232023, backend = "cmdstanr" # 选择后端 rstan 或 cmdstanr ) # 拟合结果 summary(fit_brms) # 发散很多 plot(fit_brms) # 模型诊断指标 brms::WAIC(fit_brms) brms::pp_check(fit_brms) # gee GLMMadaptive glmmML geepack 和 lme4 的模型输出结果是接近的 # blme 基于 lme4 的贝叶斯估计 fit_blme <- blme::bglmer( formula = y ~ x1 + x2 + (1 | g), data = sim_data, offset = log(o), family = poisson(link = "log") ) summary(fit_blme) # MCMCglmm 包 贝叶斯估计 fit_mcmcglmm <- MCMCglmm::MCMCglmm( fixed = y ~ x1 + x2 + offset(log(o)), random = ~g, family = "poisson", data = sim_data, verbose = FALSE ) summary(fit_mcmcglmm) -
基于 MASS 包的地形数据集 topo,建立高斯过程回归模型,比较贝叶斯预测与克里金插值预测的效果。
代码
data(topo, package = "MASS") set.seed(20232023) nchains <- 2 # 2 条迭代链 # 给每条链设置不同的参数初始值 inits_data_gaussian <- lapply(1:nchains, function(i) { list( beta = rnorm(1), sigma = runif(1), phi = runif(1), tau = runif(1) ) }) # 预测区域网格化 nx <- ny <- 27 topo_grid_df <- expand.grid( x = seq(from = 0, to = 6.5, length.out = nx), y = seq(from = 0, to = 6.5, length.out = ny) ) # 对数高斯模型 topo_gaussian_d <- list( N1 = nrow(topo), # 观测记录的条数 N2 = nrow(topo_grid_df), D = 2, # 2 维坐标 x1 = topo[, c("x", "y")], # N x 2 坐标矩阵 x2 = topo_grid_df[, c("x", "y")], y1 = topo[, "z"] # N 向量 ) library(cmdstanr) # 编码 mod_topo_gaussian <- cmdstan_model( stan_file = "code/gaussian_process_pred.stan", compile = TRUE, cpp_options = list(stan_threads = TRUE) ) # 高斯过程回归模型 fit_topo_gaussian <- mod_topo_gaussian$sample( data = topo_gaussian_d, # 观测数据 init = inits_data_gaussian, # 迭代初值 iter_warmup = 500, # 每条链预处理迭代次数 iter_sampling = 1000, # 每条链总迭代次数 chains = nchains, # 马尔科夫链的数目 parallel_chains = 2, # 指定 CPU 核心数,可以给每条链分配一个 threads_per_chain = 1, # 每条链设置一个线程 show_messages = FALSE, # 不显示迭代的中间过程 refresh = 0, # 不显示采样的进度 output_dir = "data-raw/", seed = 20232023 ) # 诊断 fit_topo_gaussian$diagnostic_summary() # 对数高斯模型 fit_topo_gaussian$summary( variables = c("lp__", "beta", "sigma", "phi", "tau"), .num_args = list(sigfig = 4, notation = "dec") ) # 未采样的位置的预测值 ypred <- fit_topo_gaussian$summary(variables = "ypred", "mean") # 预测值 topo_grid_df$ypred <- ypred$mean # 整理数据 library(sf) topo_grid_sf <- st_as_sf(topo_grid_df, coords = c("x", "y"), dim = "XY") library(stars) # 26x26 的网格 topo_grid_stars <- st_rasterize(topo_grid_sf, nx = 26, ny = 26) library(ggplot2) ggplot() + geom_stars(data = topo_grid_stars, aes(fill = ypred)) + scale_fill_viridis_c(option = "C") + theme_bw() -
用 brms 包实现贝叶斯高斯过程回归模型,考虑用样条近似高斯过程以加快计算。提示:brms 包的函数
gp()的参数 \(k\) 表示近似高斯过程 GP 所用的基函数的数目。截止写作时间,函数gp()的参数cov只能取指数二次核函数 exponentiated-quadratic kernel 。代码
# 高斯过程近似计算 bgamm2 <- brms::brm( z ~ gp(x, y, cov = "exp_quad", c = 5 / 4, k = 50), data = topo, chains = 2, seed = 20232023, warmup = 1000, iter = 2000, thin = 1, refresh = 0, control = list(adapt_delta = 0.99) ) # 输出结果 summary(bgamm2) # 条件效应 me3 <- brms::conditional_effects(bgamm1, ndraws = 200, spaghetti = TRUE) # 绘制图形 plot(me3, ask = FALSE, points = TRUE)